So far, in this blog, I’ve focused on studies related to GPT’s NLP capabilities, without really diving deep into areas where, in my opinion, GPT (or any other LLM) shouldn’t be used.
However, by using the database from my last post, I was able to spot recurring patterns in the verbs GPT uses in clinical and scientific publications.
[LINK TO THE ARTICLE ABOUT THE DATABASE]
[LINK TO THE DATABASE IN HUGGINFACE]
Sure, but how?
Well, by analyzing the cadence of verbs in the paraphrasing done by GPT of content originally written by humans in actual PubMed abstracts.
I hypothesized that the verbs used by GPT in the paraphrasing (in my database) are likely the same ones the LLM prefers to use when asked a question related to the clinical field.
Let’s quickly review the database from the previous article, this time filtering only a few rows where the value in the ‘Source’ column is ‘PubMed Abstracts’.
Column 4 contains the prompt given to the LLM to rewrite the text written by humans in Column 2. For convenience and brevity, I’ve shortened it in the table. The full prompt used is: “Rewrite and paraphrase the following sentence using your own words. Focus on changing the structure and vocabulary while preserving the original meaning and tone. Only return the rewritten sentence.”
| ID | Original Sentence | Source | Task Description | GPT Paraphrase | Original Verbs | Paraphrased Verbs |
|---|---|---|---|---|---|---|
| 10 | Eighty-six percent of these genes were downregulated by JP-8. | PubMed Abstracts | Rewrite and paraphrase the following sentence… | JP-8 led to a downregulation of eighty-six… | [‘downregulate’] | [‘lead’] |
| 18 | Surgical repair of the orbits in a patient who autoenucleated… | PubMed Abstracts | Rewrite and paraphrase the following sentence… | Surgical reconstruction of the eye sockets… | [‘autoenucleate’] | [‘have’, ‘enucleate’] |
| 33 | This review outlines the primary findings of these trials… | PubMed Abstracts | Rewrite and paraphrase the following sentence… | This review summarizes the key results of… | [‘outline’, ‘provide’] | [‘summarize’, ‘offer’] |
| 47 | This study showed the anatomic variations of LCA during… | PubMed Abstracts | Rewrite and paraphrase the following sentence… | This research revealed the anatomical… | [‘show’, ‘perform’] | [‘reveal’, ‘conduct’] |
| 65 | Four children with fecal dysfunction due to a congenital… | PubMed Abstracts | Rewrite and paraphrase the following sentence… | Interviews were conducted with four… | [‘interview’] | [‘conduct’, ‘experience’] |
Before we get into the study, let me quickly explain what Pubmed.com is and who the authors of the academic papers on this platform are (spoiler: one of the “authors” is GPT).
PubMed is a free database that primarily includes the MEDLINE database of references and abstracts on life sciences and biomedical topics. The United States National Library of Medicine (NLM) at the National Institutes of Health maintains the database as part of the Entrez system of information retrieval.
From 1971 to 1997, online access to the MEDLINE database was primarily through institutional facilities, such as university libraries. PubMed, first released in January 1996, ushered in the era of private, free, home- and office-based MEDLINE searching. The PubMed system was offered free to the public starting in June 1997.
So, in short, you can’t directly publish articles on Pubmed; it’s actually a search engine that indexes articles from scientific journals.
Now, a quick side note: what’s an abstract on Pubmed?
An abstract on PubMed is a brief summary of a scientific article published in an academic journal. It gives an overview of the key issues covered, the research methods used, the results obtained, and the conclusions drawn by the authors. The purpose of an abstract is to provide readers with a quick, concise view of the article’s content, so they can quickly determine if it’s relevant to their research or study.
But why is it important for abstracts to be written by the author, rather than by an LLM like GPT?
Because the author has a deep understanding of their own work, its nuances, and the context in which it sits. The author can accurately express the study’s intentions, results, and implications, ensuring that the summary truly captures the essence of the work and aligns with scientific standards. A model like GPT, while advanced, lacks the ability to genuinely understand the deeper meaning and implications of a specific study. This could lead to abstracts that miss the mark, misinterpret data, or overlook critical details, potentially compromising the scientific integrity of the work and misleading readers.
Alright, now that we’ve set up the environment variables, let’s dive into the juicy part of the study.
Data process
In the first part of the analysis, I extracted the verbs (only from the rows related to PubMed abstracts, as mentioned earlier) and cleaned them of stopwords (even though I had removed them during the extraction using the NLP Stanza library, I wanted to double-check). Then, I counted how many times each verb was used by both categories: human and GPT.
Next, the frequency of each verb was combined into a single dataframe, and I calculated a log base 2 ratio to see how much more often a verb was used by GPT compared to humans (or the other way around). Finally, I sorted the dataframe based on this ratio, and a chart was created for the top 50 most frequent verbs.
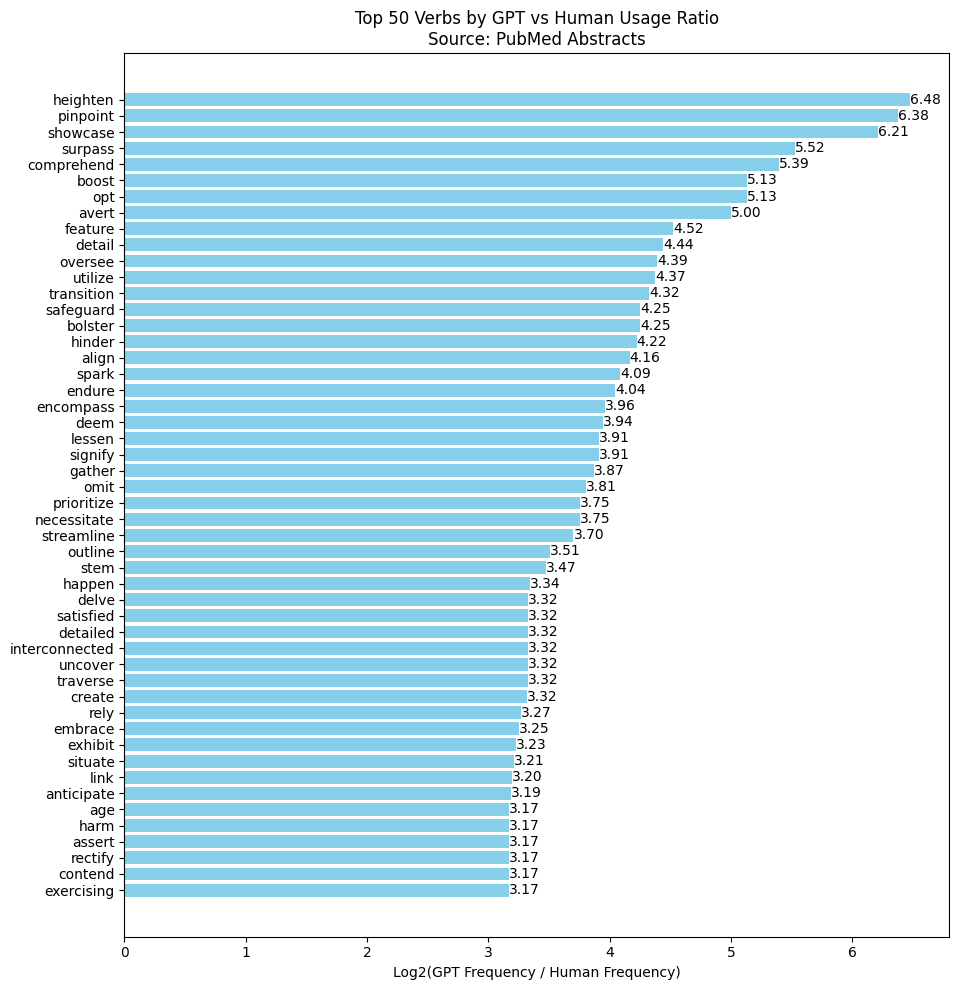
This chart, however, is somewhat incomplete: the frequency of verbs used by GPT compared to humans is only based on the database. If my goal was to identify “delve-type” verbs (verbs frequently used by GPT but rarely in everyday English), I needed an additional metric—the frequency from the Google Ngram corpus.
By adding the Ngram metric, we get a more comprehensive view of the delve-type verbs GPT uses within PubMed abstracts (see the table below for the top 10).
| Verb | Human_Frequency | GPT_Frequency | Total_Frequency | Log_Frequency_Ratio | Ngram |
| prioritize | 4 | 54 | 58 | 3.754887502 | 0.00004622472726 |
| showcase | 1 | 74 | 75 | 6.209453366 | 0.00006929563319 |
| delve | 1 | 10 | 11 | 3.321928095 | 0.00008237761866 |
| pinpoint | 1 | 83 | 84 | 6.375039431 | 0.0001081659942 |
| streamline | 1 | 13 | 14 | 3.700439718 | 0.0001205756769 |
| align | 15 | 269 | 284 | 4.164571767 | 0.0001676315991 |
| oversee | 1 | 21 | 22 | 4.392317423 | 0.0001793446549 |
| opt | 1 | 35 | 36 | 5.129283017 | 0.0002153562952 |
| interconnected | 3 | 30 | 33 | 3.321928095 | 0.000217043851 |
| uncover | 9 | 90 | 99 | 3.321928095 | 0.0002318950356 |
Now, all that remained was to verify my initial hypothesis: would GPT really use these verbs when tasked with writing abstracts for PubMed? It was time to find out.
The first step was certainly to identify how many PubMed abstracts contained those verbs over time, using the search tool that PubMed itself provides.
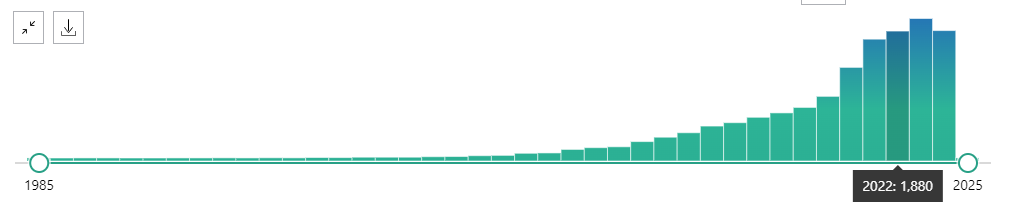
Starting from 2020-2021, the usage of the verb “Prioritize” in PubMed abstracts shows a noticeable surge, with a sharp increase in frequency. The peak occurs in 2023 with over 2500 occurrences, indicating that the verb has become much more prevalent in recent years.

Starting from 2023, the usage of the verb “Showcase” in PubMed abstracts shows a noticeable rise, with a sharp increase in frequency. The peak occurs in 2023 with over 800 occurrences, indicating that the verb has become much more common in recent years, reflecting its growing relevance in scientific and clinical discussions.
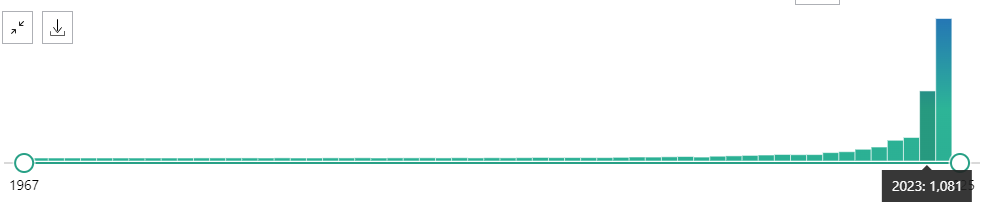
Starting from 2023, the usage of the verb “Delve” in PubMed abstracts shows a significant rise, with a sharp peak of 1,081 occurrences. This dramatic increase indicates that “Delve” has become much more prevalent in recent scientific literature, reflecting its growing use in academic and clinical contexts.
The Detector
The pieces were starting to fit together — what had initially seemed like a weak hypothesis was now yielding results. All that was left was to analyze the content of the abstracts containing the three verbs I had examined, using an AI detector. For this, I chose the most trusted one on the market, GPTZero.
GPTZero recently secured $10 million in a Series A funding round led by Footwork VC, with backing from investors like Reach Capital and former media executives. This funding is aimed at enhancing its capability to detect AI-generated content and promote responsible AI usage. As AI tools like ChatGPT become more prevalent in education, universities are increasingly incorporating GPTZero to identify AI-written submissions.
| Verb_Prioritize_in_pubmed_abstracts | % AI / Human by GptZero |
| https://pubmed.ncbi.nlm.nih.gov/39280026/ | Human almost 98% |
| https://pubmed.ncbi.nlm.nih.gov/39279808/ | over 80% by ai |
| https://pubmed.ncbi.nlm.nih.gov/39278691/ | Human 100% |
| https://pubmed.ncbi.nlm.nih.gov/39277266/ | over 70% mixed |
| https://pubmed.ncbi.nlm.nih.gov/39276987/ | over 70% mixed |
| https://pubmed.ncbi.nlm.nih.gov/39275498/ | Human almost 98% |
| https://pubmed.ncbi.nlm.nih.gov/39275195/ | Human almost 98% |
| https://pubmed.ncbi.nlm.nih.gov/39274561/ | over 80% by ai |
| https://pubmed.ncbi.nlm.nih.gov/39274494/ | over 80% by ai |
| https://pubmed.ncbi.nlm.nih.gov/39273855/ | over 80% by ai |
| Verb_Showcase_in_pubmed_abstracts | % AI / Human by GptZero |
| https://pubmed.ncbi.nlm.nih.gov/39290475/ | over 80% by ai |
| https://pubmed.ncbi.nlm.nih.gov/39290468/ | over 80% by ai |
| https://pubmed.ncbi.nlm.nih.gov/39290339/ | Human almost 98% |
| https://pubmed.ncbi.nlm.nih.gov/39289703/ | over 70% mixed |
| https://pubmed.ncbi.nlm.nih.gov/39289339/ | over 80% by ai |
| https://pubmed.ncbi.nlm.nih.gov/39288642/ | Human almost 98% |
| https://pubmed.ncbi.nlm.nih.gov/39288446/ | Almost 60% mixed |
| https://pubmed.ncbi.nlm.nih.gov/39286054/ | over 80% by ai |
| https://pubmed.ncbi.nlm.nih.gov/39285836/ | Human almost 98% |
| https://pubmed.ncbi.nlm.nih.gov/39285780/ | Almost 60% mixed |
And my favorite one:
| Verb_Delve_in_pubmed_abstracts | % AI / Human by GptZero |
| https://pubmed.ncbi.nlm.nih.gov/39291150/ | over 80% by ai |
| https://pubmed.ncbi.nlm.nih.gov/39290671/ | over 80% by ai |
| https://pubmed.ncbi.nlm.nih.gov/39290110/ | over 80% by ai |
| https://pubmed.ncbi.nlm.nih.gov/39289800/ | over 80% by ai |
| https://pubmed.ncbi.nlm.nih.gov/39288855/ | over 80% by ai |
| https://pubmed.ncbi.nlm.nih.gov/39288830/ | Human almost 98% |
| https://pubmed.ncbi.nlm.nih.gov/39288736/ | over 80% by ai |
| https://pubmed.ncbi.nlm.nih.gov/39287235/ | over 80% by ai |
| https://pubmed.ncbi.nlm.nih.gov/39286389/ | over 80% by ai |
| https://pubmed.ncbi.nlm.nih.gov/39286089/ | over 80% by ai |
Conclusion
We can now confidently say that there are specific verbs GPT tends to favor in certain contexts, and unfortunately, their presence can be a clear indicator of AI-generated text.
While AI detectors are not yet 100% reliable, I would find it alarming even if only 1% of the articles analyzed were written by AI. This raises serious concerns about the increasing involvement of AI in producing academic content and the potential implications for the credibility and authenticity of scientific research.
So, we’ve reached the end of this research. I hope you enjoyed it! If you have any questions or would like to share your thoughts about the process and tools used, feel free to email me!
This is yet another research project, another small step forward in identifying areas where AI should not be applied.